Polyploid
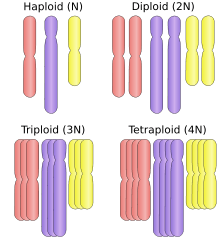
Polyploid cells and organisms are those containing more than two paired (homologous) sets of chromosomes. Most species whose cells have nuclei (Eukaryotes) are diploid, meaning they have two sets of chromosomes—one set inherited from each parent. However, polyploidy is found in some organisms and is especially common in plants. In addition, polyploidy occurs in some tissues of animals that are otherwise diploid, such as human muscle tissues.[1] This is known as endopolyploidy. Species whose cells do not have nuclei, that is, Prokaryotes, may be polyploid organisms, as seen in the large bacterium Epulopiscium fishelsoni . Hence ploidy is defined with respect to a cell. Most eukaryotes have diploid somatic cells, but produce haploid gametes (eggs and sperm) by meiosis. A monoploid has only one set of chromosomes, and the term is usually only applied to cells or organisms that are normally diploid. Male bees and other Hymenoptera, for example, are monoploid. Unlike animals, plants and multicellular algae have life cycles with two alternating multicellular generations. The gametophyte generation is haploid, and produces gametes by mitosis, the sporophyte generation is diploid and produces spores by meiosis.
Polyploidy refers to a numerical change in a whole set of chromosomes. Organisms in which a particular chromosome, or chromosome segment, is under- or overrepresented are said to be aneuploid (from the Greek words meaning "not", "good", and "fold"). Therefore, the distinction between aneuploidy and polyploidy is that aneuploidy refers to a numerical change in part of the chromosome set, whereas polyploidy refers to a numerical change in the whole set of chromosomes.[2]
Polyploidy may occur due to abnormal cell division, either during mitosis, or commonly during metaphase I in meiosis.
Polyploidy occurs in highly differentiated human tissues in the liver, heart muscle and bone marrow. It occurs in the somatic cells of some animals, such as goldfish,[3] salmon, and salamanders, but is especially common among ferns and flowering plants (see Hibiscus rosa-sinensis), including both wild and cultivated species. Wheat, for example, after millennia of hybridization and modification by humans, has strains that are diploid (two sets of chromosomes), tetraploid (four sets of chromosomes) with the common name of durum or macaroni wheat, and hexaploid (six sets of chromosomes) with the common name of bread wheat. Many agriculturally important plants of the genus Brassica are also tetraploids.
Polyploidy can be induced in plants and cell cultures by some chemicals: the best known is colchicine, which can result in chromosome doubling, though its use may have other less obvious consequences as well. Oryzalin will also double the existing chromosome content.
Types

Polyploid types are labeled according to the number of chromosome sets in the nucleus. The letter x is used to represent the number of chromosomes in a single set.
- triploid (three sets; 3x), for example seedless watermelons, common in the phylum Tardigrada[4]
- tetraploid (four sets; 4x), for example Salmonidae fish,[5] the cotton Gossypium hirsutum [6]
- pentaploid (five sets; 5x), for example Kenai Birch (Betula papyrifera var. kenaica)
- hexaploid (six sets; 6x), for example wheat, kiwifruit[7]
- heptaploid or septaploid (seven sets; 7x)
- octaploid or octoploid, (eight sets; 8x), for example Acipenser (genus of sturgeon fish), dahlias
- decaploid (ten sets; 10x), for example certain strawberries
- dodecaploid (twelve sets; 12x), for example the plant Celosia argentea or the invasive one Spartina anglica[8] or the amphibian Xenopus ruwenzoriensis.
Animals
Examples in animals are more common in non-vertebrates[9] such as flatworms, leeches, and brine shrimp. Within vertebrates, examples of stable polyploidy include the salmonids and many cyprinids (i.e. carp).[10] Some fish have as many as 400 chromosomes.[10] Polyploidy also occurs commonly in amphibians; for example the biomedically-important Xenopus genus contains many different species with as many as 12 sets of chromosomes (dodecaploid).[11] Polyploid lizards are also quite common, but are sterile and must reproduce by parthenogenesis. Polyploid mole salamanders (mostly triploids) are all female and reproduce by kleptogenesis,[12] "stealing" spermatophores from diploid males of related species to trigger egg development but not incorporating the males' DNA into the offspring. While mammalian liver cells are polyploid, rare instances of polyploid mammals are known, but most often result in prenatal death.
An octodontid rodent of Argentina's harsh desert regions, known as the plains viscacha rat (Tympanoctomys barrerae) has been reported as an exception to this 'rule'.[13] However, careful analysis using chromosome paints shows that there are only two copies of each chromosome in T. barrerae, not the four expected if it were truly a tetraploid.[14] The rodent is not a rat, but kin to guinea pigs and chinchillas. Its "new" diploid [2n] number is 102 and so its cells are roughly twice normal size. Its closest living relation is Octomys mimax, the Andean Viscacha-Rat of the same family, whose 2n = 56. It was therefore surmised that an Octomys-like ancestor produced tetraploid (i.e., 2n = 4x = 112) offspring that were, by virtue of their doubled chromosomes, reproductively isolated from their parents.
Polyploidy was induced in fish by Har Swarup (1956) using a cold-shock treatment of the eggs close to the time of fertilization, which produced triploid embryos that successfully matured.[15][16] Cold or heat shock has also been shown to result in unreduced amphibian gametes, though this occurs more commonly in eggs than in sperm.[17] John Gurdon (1958) transplanted intact nuclei from somatic cells to produce diploid eggs in the frog, Xenopus (an extension of the work of Briggs and King in 1952) that were able to develop to the tadpole stage.[18] The British Scientist, J. B. S. Haldane hailed the work for its potential medical applications and, in describing the results, became one of the first to use the word “clone” in reference to animals. Later work by Shinya Yamanaka showed how mature cells can be reprogrammed to become pluripotent, extending the possibilities to non-stem cells. Gurdon and Yamanaka were jointly awarded the Nobel Prize in 2012 for this work.[18]
Humans
True polyploidy rarely occurs in humans, although polyploid cells occur in highly differentiated tissue, such as liver parenchyma and heart muscle, and in bone marrow.[19] Aneuploidy is more common.
Polyploidy occurs in humans in the form of triploidy, with 69 chromosomes (sometimes called 69,XXX), and tetraploidy with 92 chromosomes (sometimes called 92,XXXX). Triploidy, usually due to polyspermy, occurs in about 2–3% of all human pregnancies and ~15% of miscarriages. The vast majority of triploid conceptions end as a miscarriage; those that do survive to term typically die shortly after birth. In some cases, survival past birth may extend longer if there is mixoploidy with both a diploid and a triploid cell population present.
Triploidy may be the result of either digyny (the extra haploid set is from the mother) or diandry (the extra haploid set is from the father). Diandry is mostly caused by reduplication of the paternal haploid set from a single sperm, but may also be the consequence of dispermic (two sperm) fertilization of the egg.[20] Digyny is most commonly caused by either failure of one meiotic division during oogenesis leading to a diploid oocyte or failure to extrude one polar body from the oocyte. Diandry appears to predominate among early miscarriages, while digyny predominates among triploid zygotes that survive into the fetal period. However, among early miscarriages, digyny is also more common in those cases <8.5 weeks gestational age or those in which an embryo is present. There are also two distinct phenotypes in triploid placentas and fetuses that are dependent on the origin of the extra haploid set. In digyny, there is typically an asymmetric poorly grown fetus, with marked adrenal hypoplasia and a very small placenta. In diandry, a partial hydatidiform mole develops.[20] These parent-of-origin effects reflect the effects of genomic imprinting.
Complete tetraploidy is more rarely diagnosed than triploidy, but is observed in 1–2% of early miscarriages. However, some tetraploid cells are commonly found in chromosome analysis at prenatal diagnosis and these are generally considered 'harmless'. It is not clear whether these tetraploid cells simply tend to arise during in vitro cell culture or whether they are also present in placental cells in vivo. There are, at any rate, very few clinical reports of fetuses/infants diagnosed with tetraploidy mosaicism.
Mixoploidy is quite commonly observed in human preimplantation embryos and includes haploid/diploid as well as diploid/tetraploid mixed cell populations. It is unknown whether these embryos fail to implant and are therefore rarely detected in ongoing pregnancies or if there is simply a selective process favoring the diploid cells.
Plants

Polyploidy is pervasive in plants and some estimates suggest that 30–80% of living plant species are polyploid, and many lineages show evidence of ancient polyploidy (paleopolyploidy) in their genomes.[21][22][23] Huge explosions in angiosperm species diversity appear to have coincided with the timing of ancient genome duplications shared by many species.[24] It has been established that 15% of angiosperm and 31% of fern speciation events are accompanied by ploidy increase.[25]
Polyploid plants can arise spontaneously in nature by several mechanisms, including meiotic or mitotic failures, and fusion of unreduced (2n) gametes.[26] Both autopolyploids (e.g. potato [27]) and allopolyploids (e.g. canola, wheat, cotton) can be found among both wild and domesticated plant species.
Most polyploids display novel variation or morphologies relative to their parental species, that may contribute to the processes of speciation and eco-niche exploitation.[22][26] The mechanisms leading to novel variation in newly formed allopolyploids may include gene dosage effects (resulting from more numerous copies of genome content), the reunion of divergent gene regulatory hierarchies, chromosomal rearrangements, and epigenetic remodeling, all of which affect gene content and/or expression levels.[28][29][30][31] Many of these rapid changes may contribute to reproductive isolation and speciation. However seed generated from interploidy crosses, such as between polyploids and their parent species, usually suffer from aberrant endosperm development which impairs their viability,[32][33] thus contributing to polyploid speciation.
Lomatia tasmanica is an extremely rare Tasmanian shrub that is triploid and sterile; reproduction is entirely vegetative, with all plants having the same genetic constitution.
There are few naturally occurring polyploid conifers. One example is the Coast Redwood Sequoia sempervirens, which is a hexaploid (6x) with 66 chromosomes (2n = 6x = 66), although the origin is unclear.[34]
Aquatic plants, especially the Monocotyledons, include a large number of polyploids.[35]
Crops
The induction of polyploidy is a common technique to overcome the sterility of a hybrid species during plant breeding. For example, Triticale is the hybrid of wheat (Triticum turgidum) and rye (Secale cereale). It combines sought-after characteristics of the parents, but the initial hybrids are sterile. After polyploidization, the hybrid becomes fertile and can thus be further propagated to become triticale.
In some situations, polyploid crops are preferred because they are sterile. For example, many seedless fruit varieties are seedless as a result of polyploidy. Such crops are propagated using asexual techniques, such as grafting.
Polyploidy in crop plants is most commonly induced by treating seeds with the chemical colchicine.
Examples
- Triploid crops: apple, banana, citrus, ginger, watermelon[36]
- Tetraploid crops: apple, durum or macaroni wheat, cotton, potato, canola/rapeseed, leek, tobacco, peanut, kinnow, Pelargonium
- Hexaploid crops: chrysanthemum, bread wheat, triticale, oat, kiwifruit[7]
- Octaploid crops: strawberry, dahlia, pansies, sugar cane, oca (Oxalis tuberosa)[37]
- Dodecaploid crops: some sugar cane hybrids [38]
Some crops are found in a variety of ploidies: tulips and lilies are commonly found as both diploid and triploid; daylilies (Hemerocallis cultivars) are available as either diploid or tetraploid; apples and kinnows can be diploid, triploid, or tetraploid.
Fungi
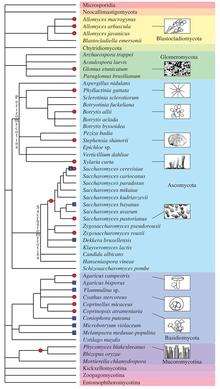
Besides plants and animals, the evolutionary history of various fungal species is dotted by past and recent whole-genome duplication events (see Albertin and Marullo 2012[39] for review). Several examples of polyploids are known:
- autopolyploid: the aquatic fungi of genus Allomyces,[40] some Saccharomyces cerevisiae strains used in bakery,[41] etc.
- allopolyploid: the widespread Cyathus stercoreus,[42] the allotetraploid lager yeast Saccharomyces pastorianus,[43] the allotriploid wine spoilage yeast Dekkera bruxellensis,[44] etc.
- paleopolyploid: the human pathogen Rhizopus oryzae,[45] the Saccharomyces genus,[46] etc.
In addition, polyploidy is frequently associated with hybridization and reticulate evolution that appear to be highly prevalent in several fungal taxa. Indeed, homoploid speciation (i.e., hybrid speciation without a change in chromosome number) has been evidenced for some fungal species (e.g., the basidiomycota Microbotryum violaceum [47]).
As for plants and animals, fungal hybrids and polyploids display structural and functional modifications compared to their progenitors and diploid counterparts. In particular, the structural and functional outcomes of polyploid Saccharomyces genomes strikingly reflect the evolutionary fate of plant polyploid ones. Large chromosomal rearrangements[48] leading to chimeric chromosomes[49] have been described, as well as more punctual genetic modifications such as gene loss.[50] The homoealleles of the allotetraploid yeast S. pastorianus show unequal contribution to the transcriptome.[51] Phenotypic diversification is also observed following polyploidization and/or hybridization in fungi,[52] producing the fuel for natural selection and subsequent adaptation and speciation.
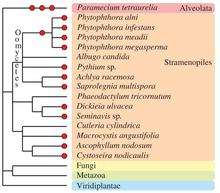
Chromalveolata
Other eukaryotic taxa have experienced one or more polyploidization events during their evolutionary history (see Albertin and Marullo, 2012[39] for review). The oomycetes, which are non-true fungi members, contain several examples of paleopolyploid and polyploid species, such as within the Phytophthora genus.[53] Some species of brown algae (Fucales, Laminariales [54] and diatoms [55]) contain apparent polyploid genomes. In the Alveolata group, the remarkable species Paramecium tetraurelia underwent three successive rounds of whole-genome duplication [56] and established itself as a major model for paleopolyploid studies.
Terminology
Autopolyploidy
Autopolyploids are polyploids with multiple chromosome sets derived from a single species. Autopolyploids can arise from a spontaneous, naturally occurring genome doubling, like the potato.[27] Others might form following fusion of 2n gametes (unreduced gametes). Bananas and apples can be found as autotriploids. Autopolyploid plants typically display polysomic inheritance, and therefore have low fertility, but may be propagated clonally.
Allopolyploidy
Allopolyploids are polyploids with chromosomes derived from different species. Precisely it is the result of multiplying the chromosome number in an F1 hybrid. Triticale is an example of an allopolyploid, having six chromosome sets, allohexaploid, four from wheat (Triticum turgidum) and two from rye (Secale cereale). Amphidiploids are a type of allopolyploids (they are allotetraploid, containing the diploid chromosome sets of both parents[57]). Some of the best examples of allopolyploids come from the Brassicas, and the Triangle of U describes the relationships between the three common diploid Brassicas (B. oleracea, B. rapa, and B. nigra) and three allotetraploids (B. napus, B. juncea, and B. carinata) derived from hybridization among the diploids.
| Species | Common Name | Family | Hybridization | Confirmed or Putative Hybridization? | Putative Parental/Introgressive species | Polyploid or Homoploid? | Polyploid Chromosome Count | References | Notes |
|---|---|---|---|---|---|---|---|---|---|
| Abelmoschus esculentus (L.) Moench | Okra | Malvaceae | Allopolyploid origin | Putative | Uncertain | Polyploid (tetraploid) | usually 2n=4x=130 | Joshi and Hardas, 1956; Schafleitner et al., 2013 | Variable ploidy |
| Actinidia deliciosa (A. Chev.) C.F.Liang & A.R.Ferguson | Kiwifruit | Actinidiaceae | Allopolyploid origin | Putative | Actinidia chinensis Planch. and Unknown | Polyploid (hexaploid) | 2n=6x=174 | Atkinson et al., 1997 | |
| Agave fourcroydes Lem. | Henequen | Asparagaceae | Allopolyploid origin | Confirmed | Uncertain | Polyploid (usu. pentaploid, triploid) | 2n=5x(3x)=150(90) | Robert et al., 2008; Hughes et al., 2007 | Variable ploidy, polyploid event not recent |
| Agave sisalana Perrine | Sisal | Asparagaceae | Allopolyploid origin | Confirmed | Uncertain | Polyploid (usu. pentaploid, hexaploid) | 2n=5x(6x)=150(180) | Robert et al., 2008 | Variable ploidy, polyploid event not recent |
| Allium ampeloprasum L. | Great headed garlic | Amaryllidaceae | Intraspecific hybrid origin | Putative | Allium ampeloprasum L. | Homoploid | - | Guenaoui et al., 2013 | |
| Allium cepa L. | Common onion | Amaryllidaceae | Interspecific hybrid origin | Putative | Uncertain: Allium vavilovii Popov & Vved., A. galanthum Kar. & Kir. or A. fistulosum L. | Homoploid | - | Gurushidze et al., 2007 | |
| Allium cornutum Clementi | Triploid onion | Amaryllidaceae | Triparental allopolyploid origin | Confirmed | Allium cepa L., A. roylei Stearn, unknown | Polyploid (triploid) | 2n=3x=24 | Fredotovic et al., 2014 | |
| Ananas comosus (L.) Merr is | Pineapple | Bromeliaceae | Interspecific introgression | Putative | Ananas ananassoides (Baker) L.B. Smith | Homoploid | - | Duval et al., 2003 | |
| Annona x atemoya | Atemoya | Annonaceae | Interspecific hybrid origin | Confirmed | Annona cherimola Mill. and A. squamosa L. | ? | - | Perfectti et al., 2004; Jalikop, 2010 | |
| Arachis hypogaea L. | Peanut | Fabaceae | Allopolyploid origin | Confirmed | Arachis duranensis Krapov. & W.C. Greg. and A. ipaënsis Krapov. & W.C. Greg. | Polypoid (tetraploid) | 2n=4x=40 | Kochert et al., 1996; Bertioli et al., 2011 | |
| Armoracia rusticana P.Gaertn., B.Mey. & Scherb. | Horseradish | Brassicaceae | Interspecific hybrid origin | Putative | Uncertain | ? | - | Courter and Rhodes, 1969 | |
| Artocarpus altilis (Parkinson ex F.A.Zorn) Fosberg | Breadfruit | Moraceae | Interspecific introgression | Putative | Artocarpus mariannensis Trécul | ? | - | Zerega et al., 2005; Jones et al., 2013 | |
| Avena sativa L. | Oat | Poaceae | Allopolyploid origin | Confirmed | Uncertain | Polyploid (hexaploid) | 2n=6x=42 | Linares et al., 1998; Oliver et al., 2013 | |
| Brassica carinata A.Braun | Ethiopian mustard | Brassicaceae | Allopolyploid origin | Confirmed | Brassica oleracea L. and B. nigra (L.) K.Koch | Polyploid (tetraploid) | 2n=4x=19 | Arias et al., 2014 | |
| Brassica juncea (L.) Czern. | Indian mustard | Brassicaceae | Allopolyploid origin | Confirmed | Brassica nigra (L.) K.Koch and B. rapa L. | Polyploid (tetraploid) | 2n=4x=18 | Arias et al., 2014 | |
| Brassica napus L. | Rapeseed, Rutabega | Brassicaceae | Allopolyploid origin | Confirmed | Brassica rapa L. and B. oleracea L. | Polyploid (tetraploid) | 2n=4x=19 | Arias et al., 2014 | |
| Cajanus cajan (L.) Millsp. | Pigeon Pea | Fabaceae | Intraspecific introgression, interspecific introgression | Putative | Wild Cajanus cajan and other species | Homoploid | - | Kassa et al., 2012 | |
| Cannabis sativa L. | Hemp | Cannabaceae | Intraspecific introgression | Putative | Cannabis sativa L. 'Indica' and 'Sativa' types | Homoploid | - | de Meijer and van Soest, 1992 | |
| Carica pentagona Heilborn | Babaco | Caricaceae | Interspecific hybrid origin | Confirmed | Uncertain (Carica stipulata V.M.Badillo, Vasconcellea pubescens A.DC., Vasconcellea weberbaueri (Harms) V.M. Badillo) | ? | - | Van Droogenbroeck et al., 2002; Van Droogenbroeck et al., 2006 | |
| Carya illinoinensis (Wangenh.) K.Koch | Pecan | Juglandaceae | Interspecific hybrid origin | Putative | Uncertain | ? | - | Grauke et al., 2011 | |
| Castanea dentata (Marshall) Borkh | Chestnut | Fagaceae | Interspecific introgression | Confirmed | Castanea pumila (L.) Mill. | Homoploid | - | Li and Dane, 2013 | Also ongoing efforts to introgress blight resistance from Castanea mollissima Blume (see Jacobs et al., 2013) |
| Castanea sativa Mill. | Chestnut | Fagaceae | Intraspecific introgression | Confirmed | Castanea sativa Eurosiberian and Mediterranean populations | Homoploid | - | Villani et al., 1999; Mattioni et al., 2013 | |
| Chenopodium quinoa Willd. | Quinoa | Chenopodiaceae | Allopolyploid origin | Putative | Uncertain | Polyploid (tetraploid) | - | Heiser, 1974; Ward, 2000; Maughan et al., 2004 | |
| Cicer arietinum L. | Chickpea (pea-shaped) | Fabaceae | Intraspecific hybrid origin | Putative | Cicer arietinum L. Desi and Kabuli Germplasm | ? | - | Upadhyaya et al., 2008; Keneni et al., 2011 | |
| Cichorium intybus L. | Radicchio | Asteraceae | Interspecific introgression | Confirmed | Wild Cichorium intybus L. | Homoploid | - | Kiaer et al., 2009 | |
| Citrus aurantiifolia (Christm.) Swingle | Key lime | Rutaceae | Interspecific hybrid origin | Confirmed | Citrus medica L. and C. subg. Papeda | ? | - | Ollitrault and Navarro, 2012; Penjor et al., 2014; Nicolosi et al., 2000; Moore, 2001 | |
| Citrus aurantium L. | Sour oranges | Rutaceae | Interspecific hybrid origin | Confirmed | Citrus maxima (Burm.) and C. reticulata Blanco | ? | - | Wu et al., 2014; Moore, 2001 | |
| Citrus clementina hort. | Clementine | Rutaceae | Interspecific hybrid origin | Confirmed | Citrus sinensis (L.) Osbeck and C. reticulata Blanco | ? | - | Wu et al., 2014 | |
| Citrus limon (L.) Osbeck | Lemon, lime | Rutaceae | Interspecific hybrid origin | Confirmed | Citrus medica L., C. aurantiifolia (Christm.) Swingle, and uncertain | ? | - | Nicolosi et al., 2000; Moore, 2001 | |
| Citrus paradisi Macfad. | Grapefruit | Rutaceae | Interspecific hybrid origin | Confirmed | Citrus sinensis (L.) Osbeck and C. maxima (Burm.) | ? | - | Wu et al., 2014; Moore, 2001 | |
| Citrus reticulata Blanco | Mandarin | Rutaceae | Interspecific introgression | Confirmed | Citrus maxima (Burm.) | ? | - | Wu et al., 2014 | |
| Citrus sinensis (L.) Osbeck | Sweet orange (blood, common) | Rutaceae | Interspecific hybrid origin | Confirmed | Uncertain | ? | - | Wu et al., 2014; Moore, 2001 | |
| Cocos nucifera L. | Coconut | Arecaceae | Intraspecific introgression | Confirmed | Cocos nucifera L. Indo-Atlantic and Pacific lineages | Homoploid | - | Gunn et al., 2011 | |
| Coffea arabica L. | Coffee | Rubiaceae | Allopolyploid origin | Confirmed | Coffea eugenioides S.Moore and C. canephora Pierre ex A.Froehner | Polyploid (tetraploid) | 2n=4x=44 | Lashermes et al., 1999 | |
| Corylus avellana L. | Hazelnut | Betulaceae | Intraspecific introgression | Confirmed | Wild Corylus avellana L. in Southern Europe | Homoploid | - | Campa et al., 2011; Boccacci et al., 2013 | |
| Cucurbita pepo L. | Winter Squash, Pumpkin | Cucurbitaceae | Intraspecific introgression | Putative | Cucurbita pepo var. texana (Scheele) D.S.Decker | Homploid | - | Kirkpatrick and Wilson, 1988 | |
| Daucus carota subsp. sativus (Hoffm.) Arcang. | Carrot | Apiaceae | Intraspecific introgression | Confirmed | Daucus carota L. subsp. carota | Homoploid | - | Iorizzo et al., 2013; Simon, 2000 | |
| Dioscorea L. spp. | Yam | Dioscoreaceae | Interspecific hybrid origin, introgression | Putative | Uncertain | Variable | - | Terauchi et al., 1992; Dansi et al., 1999; Bhattacharjee et al., 2011; Mignouna et al., 2002 | Multiple species of putative hybrid (perhaps allopolyploid) origin including Dioscorea cayennensis subsp. rotundata (Poir.) J.Miège. and D. cayennensis Lam. |
| Diospyros kaki L.f. | Persimmon | Ebenaceae | Allopolyploid origin | Putative | Uncertain | Polyploid (hexaploid) | Yonemori et al., 2008 | ||
| Ficus carica L. | Fig | Moraceae | Interspecific introgression | Putative | Uncertain | ? | - Aradhya et al., 2010 | ||
| Fragaria ananassa (Duchesne ex Weston) Duchesne ex Rozier | Strawberries | Rosaceae | Interspecific hybrid origin | Confirmed | Fragaria virginiana Mill. (octoploid), F. chiloensis (L.) Mill. (octoploid) | Homoploid relative to parentals (octoploid) | 2n=8x=56 Evans, 1977; Hirakawa et al., 2014 | Uncertain which species formed the octoploid progenitors | |
| Garcinia mangostana L. | Mangosteen | Clusiaceae | Allopolyploid origin | Putative | Garcinia celebica L. and G. malaccensis Hook.f. ex T.Anderson | Polyploid (tetraploid) | ? Richards, 1990 | Recent work shows this may not be of hybrid origin (Nazre, 2014) | |
| Gossypium hirsutum L. | Upland Cotton | Malvaceae | hybrid origin | Confirmed | Uncertain, referred to as 'A' and 'D' | Polyploid (formed <1MYA) | 2n =4x=52 Wendel and Cronn 2003 | Polyploidization likely led to agronomically significant traits (Applequist et al., 2001) | |
| Hibiscus sabdariffa L. | Roselle | Malvaceae | Allopolyploid origin | Putative | Uncertain | Polyploid (tetraploid) | 2n=4x=72 Menzel and Wilson, 1966; Satya et al., 2013 | ||
| Hordeum vulgare L. | Barley | Poaceae | Introgression | Confirmed | Hordeum spontaneum K.Koch | Homoploid | - Badr et al., 2000; Dai et al., 2012 | ||
| Humulus lupulus L. | Hops | Cannabaceae | Intraspecific introgression | Confirmed | Humulus lupulus L. North American and European Germplasm | Homoploid | - Reeves and Richards, 2011; Stajner et al., 2008; Seefelder et al., 2000 | ||
| Ipomoea batatas (L.) Lam. | Sweet Potato | Convolvulaceae | Intraspecific introgression; Interspecific introgression? | Putative | Ipomoea batatas (L.) Lam. Central American and South American Germplasm | Homoploid relative to parentals | Roullier et al., 2013 | ||
| Juglans regia L. | Walnut | Juglandaceae | Interspecific hybridization | Confirmed | Juglans sigillata Dode | Homoploid | - Gunn et al., 2010 | ||
| Lactuca sativa L. | Lettuce | Asteraceae | Intraspecific hybrid origin | Putative | Lactuca serriola L. and other L. spp. | Homoploid | - de Vries, 1997 | ||
| Lagenaria siceraria (Molina) Standl. | Bottle Gourd | Cucurbitaceae | Intraspecific introgression | Confirmed | Lagenaria siceraria (Molina) Standl. African/American and Asian Germplasm | Homoploid | - Clarke et al., 2006 | ||
| Lens culinaris Medik. ssp. culinaris | Lentil | Fabaceae | Intraspecific introgression | Putative | Wild lentil, Lens culinaris subsp. orientalis (Boiss.) Ponert | Homoploid | - Erskine et al., 2011 | ||
| Macadamia integrifolia Maiden & Betche | Macadamia | Proteaceae | Interspecific hybrid origin, interspecific introgression | Confirmed | Macadamia tetraphylla L.A.S.Johnson, and other M. spp. | Homoploid | - Hardner et al., 2009; Steiger et al., 2003; Aradhya et al., 1998 | ||
| Malus domestica Borkh. | Apple | Rosaceae | Interspecific hybrid origin | Confirmed | Malus sieversii (Ledeb.) M.Roem., M. sylvestris (L.) Mill., and possibly others | Homoploid | - Cornille et al., 2012 | ||
| Mentha piperita L. | Peppermint | Lamiaceae | Allopolyploid origin | Confirmed | Mentha aquatica L. and M. spicata L. | Polyploid (12-ploid) | 2n=12x=66 or 72 Harley and Brighton, 1977; Gobert et al., 2002 | ||
| Musa paradisiaca L. | Banana | Musaceae | Allopolyploid origin | Confirmed | Musa acuminata Colla, M. balbisiana Colla | Polyploid (usually triploid) | 2n=3x=33 Simmonds and Shepherd, 1955; Heslop-Harrison and Schwarzacher, 2007; De Langhe et al., 2010 | ||
| Nicotiana tabacum L. | Tobacco | Solanaceae | Allopolyploid origin | Confirmed | Uncertain (Nicotiana sylvestris Speg. & S. Comes and N. tomentosiformis Goodsp.) | Polyploid (tetraploid) | 2n=4x=48 Kenton et al., 1993; Murad et al., 2002 | ||
| Olea europaea L. | Olive | Oleaceae | Intraspecific introgression | Putative | Wild Olea europaea L., Eastern and Western Germplasm | Homoploid | - Kaniewski et al., 2012; Besnard et al., 2013; Breton et al., 2006; Rubio de Casas et al., 2006; Besnard et al., 2007; Besnard et al., 2000 | ||
| Opuntia L. spp. | Opuntia | Cactaceae | Interspecific hybrid origin, Allopolyploid origin | Putative | Including Opuntia ficus-indica (L.) Mill. | Polyploid, homoploid | - Hughes et al., 2007; Griffith, 2004 | ||
| Oryza sativa L. | Rice | Poaceae | Intraspecific introgression, interspecific introgression | Putative | Oryza sativa L. 'Japonica' and 'Indica' Germplasm, Oryza rufipogon Griff. | Homoploid | - Caicedo et al., 2007; Gao and Innan, 2008 | ||
| Oxalis tuberosa Molina | Oca | Oxalidaceae | Allopolyploid origin | Putative | Uncertain | Polyploid (octaploid) | 2n=8x=64 Emswhiller and Doyle, 2002; Emshwiller 2002; Emswhiller et al., 2009 | ||
| Pennisetum glaucum (L.) R.Br. | Pearl Millet | Poaceae | Intraspecific introgression | Confirmed | Wild Pennisetum glaucum (L.) R.Br. | Homoploid | - Oumar et al., 2008 | ||
| Persea americana Mill. | Avocado (Hass and other cultivars) | Lauraceae | Intraspecific introgression | Confirmed | Persea americana Mill. 'Guatamalensis', 'Drymifolia', and' Americana' | Homoploid | - Chen et al., 2008; Davis et al., 1998; Ashworth and Clegg, 2003; Douhan et al., 2011 | ||
| Phoenix dactylifera L. | Date palm | Arecaceae | Interspecific hybrid origin | Putative | Uncertain | Homoploid | - El Hadrami et al., 2011; Bennaceur et al., 1991 | ||
| Piper methysticum G.Forst. | Kava | Piperaceae | Allopolyploid origin | Putative | Piper wichmannii C. DC. and P. gibbiflorum C.DC. | Polyploid (decaploid) | 2n=10x=130 Singh, 2004; Lebot et al., 1991 | ||
| Pistacia vera L. | Pistachio | Anacardiaceae | Interspecific introgression | Putative | Pistacia atlantica Desf., P. chinensis subsp. integerrima (J. L. Stewart ex Brandis) Rech. f. | Homoploid | - Kafkas et al., 2001 | ||
| Pisum abyssinicum A.Braun | Pea | Fabaceae | Interspecific hybrid origin | Confirmed | Uncertain (Pisum fulvum Sibth. & Sm. and other P. spp.) | Homoploid | - Vershinin et al., 2003 | ||
| Pisum sativum L. | Pea | Fabaceae | Interspecific hybrid origin | Confirmed | Uncertain (Pisum sativum subsp. elatius (M.Bieb.) Asch. & Graebn. and other P. spp.) | Homoploid | - Vershinin et al., 2003 | ||
| Plinia cauliflora (Mart.) Kausel | Jaboticaba | Myrtaceae | Intraspecific hybridization, interspecific hybridization | Putative | Plinia 'Jaboticaba' and 'Cauliflora' Germplasm; P. peruviana (Poir.) Govaerts | Homoploid | - Balerdi et al., 2006 | ||
| Prunus cerasus L. | Cherry | Rosaceae | Allopolyploid origin | Confirmed | Prunus avium (L.) L. and P. fruticosa Pall. | Polyploid (tetraploid) | 2n=4x=32 Tavaud et al., 2004; Olden and Nybom, 1968 | ||
| Prunus domestica L. | Plum | Rosaceae | Allopolyploid origin | Confirmed | Uncertain (P. cerasifera Ehrh. and P. spinosa L.) | Polyploid (hexaploid) | 2n=6x=48 Zohary, 1992; Hartmann and Neumuller, 2009 | Japanese Plum is also of hybrid origin (see Hartmann and Neumuller 2009). Also hybridizes with other cultivated Prunus spp. | |
| Prunus dulcis (Mill.) D.A.Webb | Almond | Rosaceae | Interspecific introgression | Confirmed | Prunus orientalis (Mill.) Koehne and other P. spp. | Homoploid | - Delplancke et al., 2012; Delplancke et al., 2013 | ||
| Pyrus L. species | Pear | Rosaceae | Interspecific hybrid origin | Confirmed | Many species | Homoploid | - Silva et al., 2014 | Also introgression with semidomesticated populations (see Iketani et al. 2009) | |
| Raphanus raphanistrum subsp. sativus (L.) Domin | Radish | Brassicaceae | Intraspecific introgression | Confirmed | Raphanus raphanistrum L. subsp. raphanistrum | Homoploid | - Ridley et al., 2008 | ||
| Rheum L. cultivated species | Rhubarb | Polygonaceae | Interspecific hybrid origin | Putative | Unclear | Homoploid relative to parentals (tetraploid) | - Foust and Marshall, 1991; Kuhl and Deboer, 2008 | Hybrids include: Rheum rhaponticum L., R. rhabarbarum L., R. palmatum L. | |
| Rubus L. spp. | Red raspberry, Blackberry, Tayberry, Boysenberry, etc. | Rosaceae | Allopolyploid origin, interspecific hybridization | Confirmed | Many | Polyploid | - Alice et al., 2014; Alice and Campbell, 1999 | ||
| Saccharum spp. | Sugarcane | Poaceae | Allopolyploid origin | Confirmed | Saccharum officinarum L. and S. spontaneum L. | Polyploid | Variable, 2n=10-13x=100-130 Grivet et al., 1995; D'Hont et al., 1996 | ||
| Secale cereale L. | Rye | Poaceae | Interspecific hybrid origin | Confirmed | Uncertain (Secale montanum Guss., S. vavilovii Grossh.) | Homoploid | - Bartos et al., 2008; Korzun et al., 2001; Hillman, 1978; Tang et al., 2011; Salamini et al., 2002 | ||
| Sechium edule (Jacq.) Sw. | Chayote | Cucurbitaceae | Interspecific introgression | Putative | Sechium compositum (Donn. Sm.) C. Jeffrey | Homoploid | - Newstrom, 1991 | ||
| Setaria italica (L.) P.Beauv. | Foxtail millet | Poaceae | Interspecific introgression | Confirmed | Setaria viridis (L.) P.Beauv. | Homoploid | - Till-Bottraud et al., 1992 | ||
| Solanum L. spp. Section Petota | Potato | Solanaceae | Interspecific hybrid origin, allopolyploid origin, interspecific introgression | Confirmed | Including Solanum tuberosum L., S. ajanhuiri Juz. & Bukasov, S. curtilobum Juz. & Bukasov, S. juzepczukii Bukasov | Homoploid and Polyploid | - Rodriguez et al., 2010 | ||
| Solanum lycopersicum L. | Tomato | Solanaceae | Intraspecific introgression, interspecific introgression | Confirmed | Solanum lycopersicum var. cerasiforme (Dunal) D.M. Spooner, G.J. Anderson & R.K. Jansen and S. pimpinellifolium L. | Homoploid | - Blanca et al., 2012; Causse et al., 2013; Rick, 1950 | ||
| Solanum melongena L. | Eggplant | Solanaceae | Interspecific hybrid origin, interspecific introgression, intraspecific introgression | Confirmed | Solanum undatum Lam. and others; wild S. melongena L. (=S. insanum L.) | Homoploid | - Knapp et al., 2013; Meyer et al., 2012 | Hybrid origin is not confirmed, but introgression is well documented | |
| Solanum muricatum Aiton | Pepino dulce | Solanaceae | Interspecific hybrid origin, interspecific introgression | Likely | Solanum species in Series Caripensia | Homoploid | - Blanca et al., 2007 | Polyphyletic origin and extensive, ongoing introgression with wild species | |
| Spondias purpurea L. | Jocote | Anacardiaceae | Interspecific introgression | Putative | Spondias mombin L. | Homoploid | - Miller and Schaal, 2005 | ||
| Theobroma cacao L. | Cacao (Trinitario-type) | Malvaceae | Intraspecific hybrid origin | Confirmed | Theobroma cacao L. 'Forastero' and 'Criollo' Germplasm | Homoploid | - Yang et al., 2013 | ||
| Triticum aestivum L. | Bread Wheat, Spelt | Poaceae | Allopolyploid origin | Confirmed | Triticum turgidum L. (tetraploid) with Aegilops tauschii Coss. | Polyploid (hexaploid) | 2n=6x=42 Matsuoka, 2011; Dvorak, 2012 | ||
| Triticum turgidum L. | Emmer Wheat, Durum Wheat | Poaceae | Allopolyploid origin | Confirmed | Triticum urartu Thumanjan ex Gandilyan and Aegilops speltoides Tausch | Polyploid (tetraploid) | 2n=4x=28 Dvorak et al., 2012; Matsuoka, 2011; Yamane and Kawahara, 2005 | ||
| Vaccinium corymbosum L. | Highbush Blueberry | Ericaceae | Interspecific hybrid origin, interspecific introgression | Putative | Vaccinium tenellum Aiton, V. darrowii Camp, (V. virgatum Aiton, V. angustifolium Aiton) | Uncertain | - Vander Kloet, 1980; Bruederle et al., 1994; Lyrene et al., 2003; Boches et al., 2006 | Possible hybrid origin during the Plesitocene | |
| Vanilla tahitensis J.W. Moore | Tahitian vanilla | Orchidaceae | Allopolyploid origin | Confirmed | Vanilla planifolia Jacks. ex Andrews and V. odorata C.Presl | Polyploid | Variable, 2n=2x(4x)=32(64) Lubinsky et al., 2008 | ||
| Vitis rotundifolia Michx. | Grape | Vitaceae | Interspecific introgression | Confirmed | ManyVitis spp. | Homoploid | 2n=2x=38 Reisch et al., 2012; This et al., 2006 | ||
| Zea mays L. | Maize | Poaeceae | Intraspecific introgression | Confirmed | Wild Zea mays L. (teosinte, =subsp. parviglumis Iltis & Doebley) | Homoploid | - Van Heerwaarden et al., 2011; Hufford et al., 2013 |
Paleopolyploidy
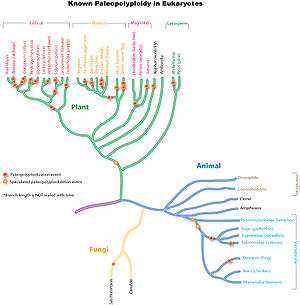
Ancient genome duplications probably occurred in the evolutionary history of all life. Duplication events that occurred long ago in the history of various evolutionary lineages can be difficult to detect because of subsequent diploidization (such that a polyploid starts to behave cytogenetically as a diploid over time) as mutations and gene translations gradually make one copy of each chromosome unlike the other copy. Over time, it is also common for duplicated copies of genes to accumulate mutations and become inactive pseudogenes.[59]
In many cases, these events can be inferred only through comparing sequenced genomes. Examples of unexpected but recently confirmed ancient genome duplications include baker's yeast (Saccharomyces cerevisiae), mustard weed/thale cress (Arabidopsis thaliana), rice (Oryza sativa), and an early evolutionary ancestor of the vertebrates (which includes the human lineage) and another near the origin of the teleost fishes. Angiosperms (flowering plants) have paleopolyploidy in their ancestry. All eukaryotes probably have experienced a polyploidy event at some point in their evolutionary history.
Karyotype
A karyotype is the characteristic chromosome complement of a eukaryote species.[60][61] The preparation and study of karyotypes is part of cytology and, more specifically, cytogenetics.
Although the replication and transcription of DNA is highly standardized in eukaryotes, the same cannot be said for their karotypes, which are highly variable between species in chromosome number and in detailed organization despite being constructed out of the same macromolecules. In some cases, there is even significant variation within species. This variation provides the basis for a range of studies in what might be called evolutionary cytology.
Paralogous
The term is used to describe the relationship between duplicated genes or portions of chromosomes that derived from a common ancestral DNA. Paralogous segments of DNA may arise spontaneously by errors during DNA replication, copy and paste transposons, or whole genome duplications.
Homologous
The term is used to describe the relationship of similar chromosomes that pair at mitosis and meiosis. In a diploid, one homolog is derived from the male parent (sperm) and one is derived from the female parent (egg). During meiosis and gametogenesis, homologous chromosomes pair and exchange genetic material by recombination, leading to the production of sperm or eggs with chromosome haplotypes containing novel genetic variation.
Homoeologous
The term homoeologous, also spelled homeologous, is used to describe the relationship of similar chromosomes or parts of chromosomes brought together following inter-species hybridization and allopolyploidization, and whose relationship was completely homologous in an ancestral species. In allopolyploids, the homologous chromosomes within each parental sub-genome should pair faithfully during meiosis, leading to disomic inheritance; however in some allopolyploids, the homoeologous chromosomes of the parental genomes may be nearly as similar to one another as the homologous chromosomes, leading to tetrasomic inheritance (four chromosomes pairing at meiosis), intergenomic recombination, and reduced fertility.
Example of homoeologous chromosomes
Durum wheat is the result of the inter-species hybridization of two diploid grass species Triticum urartu and Aegilops speltoides. Both diploid ancestors had two sets of 7 chromosomes, which were similar in terms of size and genes contained on them. Durum wheat contains two sets of chromosomes derived from Triticum urartu and two sets of chromosomes derived from Aegilops speltoides. Each chromosome pair derived from the Triticum urartu parent is homoeologous to the opposite chromosome pair derived from the Aegilops speltoides parent, though each chromosome pair unto itself is homologous.
Polyploidization and speciation
Polyploidization is a mechanism of sympatric speciation because polyploids are usually unable to interbreed with their diploid ancestors. An example is the plant Mimulus peregrinus. Sequencing confirmed that this species originated from M. x robertsii, a sterile triploid hybrid between M. guttatus and M. luteus, both of which have been introduced and naturalised in the United Kingdom. New populations of M. peregrinus arose on the Scottish mainland and the Orkney Islands via genome duplication from local populations of M. x robertsii.[62]
See also
- Klerokinesis, a type of cell division that occurs after mitosis is complete, restoring a diploid chromosome number
- Polyploid complex
- Polysomy
- Sympatry
References
- ↑ Parmacek, Michael S.; Epstein, Jonathan A. (2009). "Cardiomyocyte Renewal". New England Journal of Medicine. 361 (1): 86–8. doi:10.1056/NEJMcibr0903347. PMID 19571289.
- ↑ Griffiths, Anthony J. F. (1999). An Introduction to genetic analysis. San Francisco: W.H. Freeman. ISBN 0-7167-3520-2.
- ↑ Ohno, Susumu; Muramoto, Junichi; Christian, Lawrence; Atkin, Niels B. (1967). "Diploid-tetraploid relationship among old-world members of the fish family Cyprinidae". Chromosoma. 23 (1): 1–9. doi:10.1007/BF00293307.
- ↑ Bertolani R (2001). "Evolution of the reproductive mechanisms in Tardigrades: a review". Zoologischer Anzeiger. 240 (3–4): 247–252. doi:10.1078/0044-5231-00032.
- ↑ Stouder, Deanna J.; Bisson, Peter A.; Naiman, Robert J. (1997). Pacific Salmon and Their Ecosystems: Status and Future Options. Springer. pp. 30–1. ISBN 978-0-412-98691-8. Retrieved 9 July 2013.
- ↑ Adams, Keith L; Wendel, Jonathan F (2005). "Polyploidy and genome evolution in plants". Current Opinion in Plant Biology. 8 (2): 135–41. doi:10.1016/j.pbi.2005.01.001. PMID 15752992.
- 1 2 Crowhurst, Ross N.; Whittaker, D.; Gardner, R. C. "The genetic origin of kiwifruit".
- ↑ Ainouche, M. L.; Fortune, P. M.; Salmon, A.; Parisod, C.; Grandbastien, M.-A.; Fukunaga, K.; Ricou, M.; Misset, M.-T. (2008). "Hybridization, polyploidy and invasion: Lessons from Spartina (Poaceae)". Biological Invasions. 11 (5): 1159–73. doi:10.1007/s10530-008-9383-2.
- ↑ Otto, Sarah P; Whitton, Jeannette (2000). "Polyploidincidence Andevolution". Annual Review of Genetics. 34: 401–437. doi:10.1146/annurev.genet.34.1.401. PMID 11092833.
- 1 2 Leggatt, Rosalind A.; Iwama, George K. (2003). "Occurrence of polyploidy in the fishes". Reviews in Fish Biology and Fisheries. 13 (3): 237–46. doi:10.1023/B:RFBF.0000033049.00668.fe.
- ↑ Cannatella, David C.; De Sa, Rafael O. (1993). "Xenopus laevis as a Model Organism". Society of Systematic Biologists. 42 (4): 476–507. doi:10.1093/sysbio/42.4.476.
- ↑ Bonen, L.; Bi, James P.; Fu, Ke; Noble, Jinzong; Niedzwiecki, Daniel W.A.; Niedzwiecki, John (2007). "Unisexual salamanders (genus Ambystoma) present a new reproductive mode for eukaryotes". Genome. 50 (2): 119–36. doi:10.1139/g06-152. PMID 17546077.
- ↑ Gallardo, M.H.; González, C.A.; Cebrián, I. (2006). "Molecular cytogenetics and allotetraploidy in the red vizcacha rat, Tympanoctomys barrerae (Rodentia, Octodontidae)". Genomics. 88 (2): 214–21. doi:10.1016/j.ygeno.2006.02.010. PMID 16580173.
- ↑ Svartman, Marta; Stone, Gary; Stanyon, Roscoe (2005). "Molecular cytogenetics discards polyploidy in mammals". Genomics. 85 (4): 425–30. doi:10.1016/j.ygeno.2004.12.004. PMID 15780745.
- ↑ Swarup, H. (1956). "Production of Heteroploidy in the Three-Spined Stickleback, Gasterosteus aculeatus (L.)". Nature. 178 (4542): 1124–1125. Bibcode:1956Natur.178.1124S. doi:10.1038/1781124a0.
- ↑ Swarup, H. (1959). "Production of triploidy ingasterosteus aculeatus (L)". Journal of Genetics. 56 (2): 129–142. doi:10.1007/BF02984740.
- ↑ Mable, B.K.; Alexandrou, M. A.; Taylor, M. I. (2011). "Genome duplication in amphibians and fish: an extended synthesis". Journal of Zoology. 284: 151–182. doi:10.1111/j.1469-7998.2011.00829 (inactive 2015-02-01).
- 1 2 "Nobel Prize in Physiology or Medicine 2012 Awarded for Discovery That Mature Cells Can Be Reprogrammed to Become Pluripotent". ScienceDaily. 8 Oct 2012.
- ↑ Winkelmann, M; Pfitzer, P; Schneider, W (1987). "Significance of polyploidy in megakaryocytes and other cells in health and tumor disease". Klinische Wochenschrift. 65 (23): 1115–31. doi:10.1007/BF01734832. PMID 3323647.
- 1 2 Baker, Phil; Monga, Ash; Baker, Philip (2006). Gynaecology by ten teachers. London: Arnold. ISBN 0-340-81662-7.
- ↑ Meyers, Lauren Ancel; Levin, Donald A. (2006). "On the Abundance of Polyploids in Flowering Plants". Evolution. 60 (6): 1198–206. doi:10.1111/j.0014-3820.2006.tb01198.x. PMID 16892970.
- 1 2 Rieseberg, L. H.; Willis, J. H. (2007). "Plant Speciation". Science. 317 (5840): 910–4. Bibcode:2007Sci...317..910R. doi:10.1126/science.1137729. PMC 2442920
 . PMID 17702935.
. PMID 17702935. - ↑ Otto, Sarah P. (2007). "The Evolutionary Consequences of Polyploidy". Cell. 131 (3): 452–62. doi:10.1016/j.cell.2007.10.022. PMID 17981114.
- ↑ Debodt, S; Maere, S; Vandepeer, Y (2005). "Genome duplication and the origin of angiosperms". Trends in Ecology & Evolution. 20 (11): 591–7. doi:10.1016/j.tree.2005.07.008. PMID 16701441.
- ↑ Wood, T. E.; Takebayashi, N.; Barker, M. S.; Mayrose, I.; Greenspoon, P. B.; Rieseberg, L. H. (2009). "The frequency of polyploid speciation in vascular plants". Proceedings of the National Academy of Sciences. 106 (33): 13875–9. Bibcode:2009PNAS..10613875W. doi:10.1073/pnas.0811575106. JSTOR 40484335. PMC 2728988
 . PMID 19667210.
. PMID 19667210. - 1 2 Comai, Luca (2005). "The advantages and disadvantages of being polyploid". Nature Reviews Genetics. 6 (11): 836–46. doi:10.1038/nrg1711. PMID 16304599.
- 1 2 Xu, Xun; Xu, Shengkai; Pan, Shifeng; Cheng, Bo; Zhang, Desheng; Mu, Peixiang; Ni, Gengyun; Zhang, Shuang; Yang, Ruiqiang; Li, Jun; Wang, Gisella; Orjeda, Frank; Guzman, Michael; Torres, Roberto; Lozano, Olga; Ponce, Diana; Martinez, Germán; De La Cruz, S. K.; Chakrabarti, Virupaksh U.; Patil, Konstantin G.; Skryabin, Boris B.; Kuznetsov, Nikolai V.; Ravin, Tatjana V.; Kolganova, Alexey V.; Beletsky, Andrei V.; Mardanov, Alex; Di Genova, Daniel M.; Bolser, David M. A.; Martin, Guangcun; Li, Yu (2011). "Genome sequence and analysis of the tuber crop potato". Nature. 475 (7355): 189–95. doi:10.1038/nature10158. PMID 21743474.
- ↑ Osborn, Thomas C.; Pires, J.; Birchler, James A.; Auger, Donald L.; Chen, Z.; Lee, Hyeon-Se; Comai, Luca; Madlung, Andreas; Doerge, R.W.; Colot, Vincent; Martienssen, Robert A. (2003). "Understanding mechanisms of novel gene expression in polyploids". Trends in Genetics. 19 (3): 141–7. doi:10.1016/S0168-9525(03)00015-5. PMID 12615008.
- ↑ Chen, Z. Jeffrey; Ni, Zhongfu (2006). "Mechanisms of genomic rearrangements and gene expression changes in plant polyploids". BioEssays. 28 (3): 240–52. doi:10.1002/bies.20374. PMC 1986666
 . PMID 16479580.
. PMID 16479580. - ↑ Chen, Z. Jeffrey (2007). "Genetic and Epigenetic Mechanisms for Gene Expression and Phenotypic Variation in Plant Polyploids". Annual Review of Plant Biology. 58: 377–406. doi:10.1146/annurev.arplant.58.032806.103835. PMC 1949485
 . PMID 17280525.
. PMID 17280525. - ↑ Albertin, W.; Balliau, T; Brabant, P; Chèvre, AM; Eber, F; Malosse, C; Thiellement, H (2006). "Numerous and Rapid Nonstochastic Modifications of Gene Products in Newly Synthesized Brassica napus Allotetraploids". Genetics. 173 (2): 1101–13. doi:10.1534/genetics.106.057554. PMC 1526534
 . PMID 16624896.
. PMID 16624896. - ↑ Pennington, PD; Costa, LM; Gutierrez-Marcos, JF; Greenland, AJ; Dickinson, HG (Apr 2008). "When genomes collide: aberrant seed development following maize interploidy crosses.". Annals of Botany. 101 (6): 833–43. doi:10.1093/aob/mcn017. PMID 18276791.
- ↑ von Wangenheim, Karl-Hartmut; Peterson, Hans-Peter (2004). "Aberrant endosperm development in interploidy crosses reveals a timer of differentiation". Developmental Biology. 270 (2): 277–289. doi:10.1016/j.ydbio.2004.03.014. PMID 15183714.
- ↑ Ahuja MR, Neale DB (2002). "Origins of Polyploidy in Coast Redwood (Sequoia sempervirens (D. DON) ENDL.) and Relationship of Coast Redwood to other Genera of Taxodiaceae". Silvae Genetica. 51: 2–3.
- ↑ Les, D.H.; Philbrick, C.T. (1993). "Studies of hybridization and chromosome number variation in aquatic angiosperms: Evolutionary implications". Aquatic Botany. 44 (2–3): 181–228. doi:10.1016/0304-3770(93)90071-4.
- ↑ Seedless Fruits Make Others Needless
- ↑ Emshwiller, E. (2006). "Origins of polyploid crops: The example of the octaploid tuber crop Oxalis tuberosa". In M.A. Zeder; D. Decker-Walters; E. Emshwiller; D. Bradley; B.D. Smith. Documenting domestication: new genetic and archaeological paradigms. Berkeley, USA: University of California Press. pp. 153–168.
- ↑ Cunff; et al. (2008). "Diploid/Polyploid Syntenic Shuttle Mapping and Haplotype-Specific Chromosome Walking Toward a Rust Resistance Gene (Bru1) in Highly Polyploid Sugarcane (2n ∼ 12x ∼ 115)".
- 1 2 3 4 Albertin, W.; Marullo, P. (2012). "Polyploidy in fungi: Evolution after whole-genome duplication". Proceedings of the Royal Society B. 279 (1738): 2497–509. doi:10.1098/rspb.2012.0434. PMC 3350714
 . PMID 22492065.
. PMID 22492065. - ↑ Emerson, Ralph; Wilson, Charles M. (1954). "Interspecific Hybrids and the Cytogenetics and Cytotaxonomy of Euallomyces". Mycologia. 46 (4): 393–434. JSTOR 4547843.
- ↑ Albertin, W.; Marullo, P.; Aigle, M.; Bourgais, A.; Bely, M.; Dillmann, C.; De Vienne, D.; Sicard, D. (2009). "Evidence for autotetraploidy associated with reproductive isolation inSaccharomyces cerevisiae: Towards a new domesticated species". Journal of Evolutionary Biology. 22 (11): 2157–70. doi:10.1111/j.1420-9101.2009.01828.x. PMID 19765175.
- ↑ Lu, Benjamin C. (1964). "Polyploidy in the Basidiomycete Cyathus stercoreus". American Journal of Botany. 51 (3): 343–7. doi:10.2307/2440307. JSTOR 2440307.
- ↑ Libkind, D.; Hittinger, C. T.; Valerio, E.; Goncalves, C.; Dover, J.; Johnston, M.; Goncalves, P.; Sampaio, J. P. (2011). "Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast". Proceedings of the National Academy of Sciences. 108 (35): 14539–44. Bibcode:2011PNAS..10814539L. doi:10.1073/pnas.1105430108. PMC 3167505
 . PMID 21873232.
. PMID 21873232. - ↑ Borneman Anthony R.; Zeppel Ryan; Chambers Paul J.; Curtin Chris D. (2014). "Insights into the Dekkera bruxellensis Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates". PLoS Genet. 10: e1004161. doi:10.1371/journal.pgen.1004161.
- ↑ Ma, Li-Jun; Ibrahim, Ashraf S.; Skory, Christopher; Grabherr, Manfred G.; Burger, Gertraud; Butler, Margi; Elias, Marek; Idnurm, Alexander; Lang, B. Franz; Sone, Teruo; Abe, Ayumi; Calvo, Sarah E.; Corrochano, Luis M.; Engels, Reinhard; Fu, Jianmin; Hansberg, Wilhelm; Kim, Jung-Mi; Kodira, Chinnappa D.; Koehrsen, Michael J.; Liu, Bo; Miranda-Saavedra, Diego; O'Leary, Sinead; Ortiz-Castellanos, Lucila; Poulter, Russell; Rodriguez-Romero, Julio; Ruiz-Herrera, José; Shen, Yao-Qing; Zeng, Qiandong; Galagan, James; Birren, Bruce W. (2009). Madhani, Hiten D, ed. "Genomic Analysis of the Basal Lineage Fungus Rhizopus oryzae Reveals a Whole-Genome Duplication". PLoS Genetics. 5 (7): e1000549. doi:10.1371/journal.pgen.1000549. PMC 2699053
 . PMID 19578406.
. PMID 19578406. - ↑ Wong, S.; Butler, G.; Wolfe, K. H. (2002). "Gene order evolution and paleopolyploidy in hemiascomycete yeasts". Proceedings of the National Academy of Sciences. 99 (14): 9272–7. Bibcode:2002PNAS...99.9272W. doi:10.1073/pnas.142101099. JSTOR 3059188. PMC 123130
 . PMID 12093907.
. PMID 12093907. - ↑ Devier, B.; Aguileta, G.; Hood, M. E.; Giraud, T. (2009). "Using phylogenies of pheromone receptor genes in the Microbotryum violaceum species complex to investigate possible speciation by hybridization". Mycologia. 102 (3): 689–96. doi:10.3852/09-192. PMID 20524600.
- ↑ Dunn, B.; Sherlock, G. (2008). "Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus". Genome Research. 18 (10): 1610–23. doi:10.1101/gr.076075.108. PMC 2556262
 . PMID 18787083.
. PMID 18787083. - ↑ Nakao, Y.; Kanamori, T.; Itoh, T.; Kodama, Y.; Rainieri, S.; Nakamura, N.; Shimonaga, T.; Hattori, M.; Ashikari, T. (2009). "Genome Sequence of the Lager Brewing Yeast, an Interspecies Hybrid". DNA Research. 16 (2): 115–29. doi:10.1093/dnares/dsp003. PMC 2673734
 . PMID 19261625.
. PMID 19261625. - ↑ Scannell, Devin R.; Byrne, Kevin P.; Gordon, Jonathan L.; Wong, Simon; Wolfe, Kenneth H. (2006). "Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts". Nature. 440 (7082): 341–5. Bibcode:2006Natur.440..341S. doi:10.1038/nature04562. PMID 16541074.
- ↑ Minato, Toshiko; Yoshida, Satoshi; Ishiguro, Tatsuji; Shimada, Emiko; Mizutani, Satoru; Kobayashi, Osamu; Yoshimoto, Hiroyuki (2009). "Expression profiling of the bottom fermenting yeastSaccharomyces pastorianusorthologous genes using oligonucleotide microarrays". Yeast. 26 (3): 147–65. doi:10.1002/yea.1654. PMID 19243081.
- ↑ Lidzbarsky, Gabriel A.; Shkolnik, Tamar; Nevo, Eviatar (2009). Idnurm, Alexander, ed. "Adaptive Response to DNA-Damaging Agents in Natural Saccharomyces cerevisiae Populations from "Evolution Canyon", Mt. Carmel, Israel". PLoS ONE. 4 (6): e5914. Bibcode:2009PLoSO...4.5914L. doi:10.1371/journal.pone.0005914. PMC 2690839
 . PMID 19526052.
. PMID 19526052. - ↑ Ioos, Renaud; Andrieux, Axelle; Marçais, Benoît; Frey, Pascal (2006). "Genetic characterization of the natural hybrid species Phytophthora alni as inferred from nuclear and mitochondrial DNA analyses". Fungal Genetics and Biology. 43 (7): 511–29. doi:10.1016/j.fgb.2006.02.006. PMID 16626980.
- ↑ Phillips, N.; Kapraun, D. F.; Gómez Garreta, A.; Ribera Siguan, M. A.; Rull, J. L.; Salvador Soler, N.; Lewis, R.; Kawai, H. (2011). "Estimates of nuclear DNA content in 98 species of brown algae (Phaeophyta)". AoB Plants. 2011: plr001. doi:10.1093/aobpla/plr001. PMC 3064507
 . PMID 22476472.
. PMID 22476472. - ↑ Chepurnov, Victor A.; Mann, David G.; Vyverman, Wim; Sabbe, Koen; Danielidis, Daniel B. (2002). "Sexual Reproduction, Mating System, and Protoplast Dynamics of Seminavis (Bacillariophyceae)". Journal of Phycology. 38 (5): 1004–19. doi:10.1046/j.1529-8817.2002.t01-1-01233.x.
- ↑ Aury, Jean-Marc; Jaillon, Olivier; Duret, Laurent; Noel, Benjamin; Jubin, Claire; Porcel, Betina M.; Ségurens, Béatrice; Daubin, Vincent; Anthouard, Véronique; Aiach, Nathalie; Arnaiz, Olivier; Billaut, Alain; Beisson, Janine; Blanc, Isabelle; Bouhouche, Khaled; Câmara, Francisco; Duharcourt, Sandra; Guigo, Roderic; Gogendeau, Delphine; Katinka, Michael; Keller, Anne-Marie; Kissmehl, Roland; Klotz, Catherine; Koll, France; Le Mouël, Anne; Lepère, Gersende; Malinsky, Sophie; Nowacki, Mariusz; Nowak, Jacek K.; et al. (2006). "Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia". Nature. 444 (7116): 171–8. Bibcode:2006Natur.444..171A. doi:10.1038/nature05230. PMID 17086204.
- ↑ Rieger, R.; Michaelis, A.; Green, M.M. (1968). A glossary of genetics and cytogenetics: Classical and molecular.
- ↑ Warschefsky, E.; Penmetsa, R. V.; Cook, D. R.; von Wettberg, E. J. B. (8 October 2014). "Back to the wilds: Tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives". American Journal of Botany. 101 (10): 1791–1800. doi:10.3732/ajb.1400116. PMID 25326621.
- ↑ Edger, Patrick P.; Pires, Chris J. (2009). "Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes". Chromosome Research. 17 (5): 699–717. doi:10.1007/s10577-009-9055-9. PMID 19802709.
- ↑ White M.J.D. 1973. The chromosomes. 6th ed, Chapman & Hall, London. p28
- ↑ Stebbins G.L. 1950. Variation and evolution in plants. Chapter XII: The Karyotype. Columbia University Press N.Y.
- ↑ Vallejo-Marín Mario; Buggs Richard J. A.; Cooley Arielle M.; Puzey Joshua R. (2015). "Speciation by genome duplication: Repeated origins and genomic composition of the recently formed allopolyploid species Mimulus peregrinus". Evolution. 69: 1487–1500. doi:10.1111/evo.12678.
Further reading
- Snustad, D. Peter; et al. (2006). Principles of Genetics (4th ed.). Hoboken, NJ: John Wiley & Sons. ISBN 0-471-69939-X.
- The Arabidopsis Genome Initiative (2000). "Analysis of the genome sequence of the flowering plant Arabidopsis thaliana". Nature. 408 (6814): 796–815. doi:10.1038/35048692. PMID 11130711.
- Eakin, Guy S.; Behringer, Richard R. (2003). "Tetraploid development in the mouse". Developmental Dynamics. 228 (4): 751–66. doi:10.1002/dvdy.10363. PMID 14648853.
- Gaeta, R. T.; Pires, J. C.; Iniguez-Luy, F.; Leon, E.; Osborn, T. C. (2007). "Genomic Changes in Resynthesized Brassica napus and Their Effect on Gene Expression and Phenotype". The Plant Cell Online. 19 (11): 3403–3417. doi:10.1105/tpc.107.054346. PMC 2174891
 . PMID 18024568.
. PMID 18024568. - Gregory, T.R.; Mable, B.K. (2005). "Polyploidy in animals". In Gregory, T.R. The Evolution of the Genome. San Diego: Elsevier. pp. 427–517.
- Jaillon, Olivier; Aury, Jean-Marc; Brunet, Frédéric; Petit, Jean-Louis; et al. (2004). "Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype". Nature. 431 (7011): 946–57. Bibcode:2004Natur.431..946J. doi:10.1038/nature03025. PMID 15496914.
- Paterson, Andrew H.; Bowers, John E.; Van De Peer, Yves; Vandepoele, Klaas (2005). "Ancient duplication of cereal genomes". New Phytologist. 165 (3): 658–61. doi:10.1111/j.1469-8137.2005.01347.x. PMID 15720677.
- Raes, Jeroen; Vandepoele, Klaas; Simillion, Cedric; Saeys, Yvan; Van De Peer, Yves (2003). "Investigating ancient duplication events in the Arabidopsis genome". Journal of Structural and Functional Genomics. 3 (1–4): 117–29. doi:10.1023/A:1022666020026. PMID 12836691.
- Simillion, C.; Vandepoele, K; Van Montagu, MC; Zabeau, M; Van De Peer, Y (2002). "The hidden duplication past of Arabidopsis thaliana". Proceedings of the National Academy of Sciences. 99 (21): 13627–32. Bibcode:2002PNAS...9913627S. doi:10.1073/pnas.212522399. JSTOR 3073458. PMC 129725
 . PMID 12374856.
. PMID 12374856. - Soltis, Douglas E.; Soltis, Pamela S.; Schemske, Douglas W.; Hancock, James F.; Thompson, John N.; Husband, Brian C.; Judd, Walter S. (2007). "Autopolyploidy in Angiosperms: Have We Grossly Underestimated the Number of Species?". Taxon. 56 (1): 13–30. JSTOR 25065732.
- Soltis DE, Buggs RJA, Doyle JJ, Soltis PS (2010). "What we still don't know about polyploidy". Taxon. 59: 1387–403. JSTOR 20774036.
- Taylor, J. S.; Braasch, I; Frickey, T; Meyer, A; Van De Peer, Y (2003). "Genome Duplication, a Trait Shared by 22,000 Species of Ray-Finned Fish". Genome Research. 13 (3): 382–90. doi:10.1101/gr.640303. PMC 430266
 . PMID 12618368.
. PMID 12618368. - Tate, J.A.; Soltis, D.E.; Soltis, P.S. (2005). "Polyploidy in plants". In Gregory, T.R. The Evolution of the Genome. San Diego: Elsevier. pp. 371–426.
- Van De Peer, Yves; Taylor, John S.; Meyer, Axel (2003). "Are all fishes ancient polyploids?". Journal of Structural and Functional Genomics. 3 (1–4): 65–73. doi:10.1023/A:1022652814749. PMID 12836686.
- Van De Peer, Yves (2004). "Tetraodon genome confirms Takifugu findings: Most fish are ancient polyploids". Genome Biology. 5 (12): 250. doi:10.1186/gb-2004-5-12-250. PMC 545788
 . PMID 15575976.
. PMID 15575976. - Van de Peer, Y.; Meyer, A. (2005). "Large-scale gene and ancient genome duplications". In Gregory, T.R. The Evolution of the Genome. San Diego: Elsevier. pp. 329–68.
- Wolfe, Kenneth H.; Shields, Denis C. (1997). "Molecular evidence for an ancient duplication of the entire yeast genome". Nature. 387 (6634): 708–13. doi:10.1038/42711. PMID 9192896.
- Wolfe, Kenneth H. (2001). "Yesterday's polyploids and the mystery of diploidization". Nature Reviews Genetics. 2 (5): 333–41. doi:10.1038/35072009. PMID 11331899.
External links
- Polyploidy on Kimball's Biology Pages
- The polyploidy portal a community-editable project with information, research, education, and a bibliography about polyploidy.