Unequal crossing over
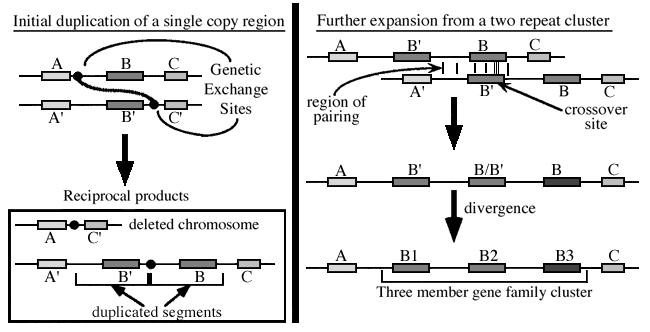
Unequal crossing over is a type of gene duplication or deletion event that deletes a sequence in one strand and replaces it with a duplication from its sister chromatid in mitosis or from its homologous chromosome during meiosis. It is a type of chromosomal crossover between homologous sequences that are not paired precisely. Normally genes are responsible for occurrence of crossing over. It exchanges sequences of different links between chromosomes. Along with gene conversion, it is believed to be the main driver for the generation of gene duplications and is a source of mutation in the genome.[1]
Mechanisms
During meiosis, the duplicated chromosomes in eukaryotic organisms attach to each other in the centromere region and pair to each other (chromatids). The maternal and paternal chromosomes then align alongside each other. During this time, recombination can take place via crossing over of sections of the paternal and maternal chromatids and leads to reciprocal recombination or non-reciprocal recombination.[1] Unequal crossing over requires a measure of similarity between the sequences for misalignment to occur. The more similarity within the sequences, the more likely unequal crossing over will occur.[1] One of the sequences is thus lost and replaced with the duplication of another sequence.
When two sequences are misaligned, unequal crossing over may create a tandem repeat on one chromosome and a deletion on the other. The rate of unequal crossing over will increase with the number of repeated sequences around the duplication. This is because these repeated sequences will pair together, allowing for the mismatch in the cross over point to occur.[2]
Consequences for the organism
Unequal crossing over is the process most responsible for creating regional gene duplications in the genome.[1] Repeated rounds of unequal crossing over cause the homogenization of the two sequences. With the increase in the duplicates, unequal crossing over can lead to dosage imbalance in the genome and can be highly deleterious.[1][2]
Evolutionary implications
In unequal crossing over, there can be large sequence exchanges between the chromosomes. Compared with gene conversion, which can only transfer a maximum of 1,500 base pairs, unequal crossing over in yeast rDNA genes has been found to transfer about 20,000 base pairs in a single crossover event[1][3] Unequal crossover can be followed by the concerted evolution of duplicated sequences.
It has been suggested that longer intron found between two beta-globin genes are a response to deleterious selection from unequal crossing over in the beta-globin genes.[1][4] Comparisons between alpha-globin, which does not have long introns, and beta-globin genes show that alpha-globin have 50 times higher concerted evolution.
When unequal crossing over creates a gene duplication, the duplicate has 4 evolutionary fates. This is due the fact that purifying selection acting on a duplicated copy is not very strong. Now that there is a redundant copy, neutral mutations can act on the duplicate. Most commonly the neutral mutations will continue until the duplicate becomes a pseudogene. If the duplicate copy increases the dosage effect of the gene product, then the duplicate may be retained as a redundant copy. Neofunctionalization is also a possibility: the duplicated copy acquires a mutation that gives it a different function than its ancestor. If both copies acquire mutations, it is possible that a subfunctional event occurs. This happens when both of the duplicated sequences have a more specialized function than the ancestral copy[5]
Genome size
Gene duplications are the main reason for the increase of genome size, and as unequal crossing over is the main mechanism for gene duplication, unequal crossing over contributes to genome size evolution is the most common regional duplication event that increases the size of the genome.
Junk DNA
When viewing the genome of a eukaryote, a striking observation is the large amount of tandem, repetitive DNA sequences that make up a large portion of the genome. For example, over 50% of the Dipodmys ordii genome is made up of three specific repeats. Drosophila virilis has three sequences that make up 40% of the genome, and 35% of the Absidia glauca is repetitive DNA sequences.[1] These short sequences have no selection pressure acting on them and the frequency of the repeats can be changed by unequal crossing over.[6]
References
- 1 2 3 4 5 6 7 8 Graur, Dan; Li, Wen-Hsiung (2000). Fundamentals of Molecular Evolution (Second ed.). Sunderland, Massachusetts: Sinauer Associates, Inc. ISBN 0878932666.
- 1 2 Russel, Peter J. (2002). iGenetics. San Francisco: Benjamin Cummings. ISBN 0-8053-4553-1.
- ↑ Szostak, J. W.; Wu, R. (1980). "Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae". Nature. 284 (5755): 426–430. doi:10.1038/284426a0.
- ↑ Zimmer, E. A.; Martin, S. M.; Beverley, S. M.; Kan, Y. W.; Wilson, A. C. (1980). "Rapid duplication and loss of genes coding for the alpha chains of hemoglobin". Proc. Natl. Acad. Sci. USA. 77: 2158–2162. doi:10.1073/pnas.77.4.2158. PMC 348671
 . PMID 6929543.
. PMID 6929543. - ↑ Force, Allan; Lynch, Michael; Pickettb, F. Bryan; Amoresa, Angel; Yana, Yi-lin; Postlethwaita, John (1999). "Preservation of Duplicate Genes by Complementary, Degenerative Mutations". Genetics. 151 (4): 1531–1545. PMC 1460548
 . PMID 10101175.
. PMID 10101175. - ↑ Zhang, J. (2003). "Evolution of the human ASPM gene, a major determinant of brain size". Genetics. 165 (4): 2063–2070. PMC 1462882
 . PMID 14704186.
. PMID 14704186.