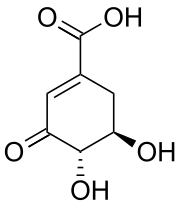
3-Dehydroshikimic acid
 | |
| Names | |
|---|---|
| IUPAC name
(4S,5R)-4,5-Ddihydroxy-3-oxocyclohexene-1-carboxylic acid | |
| Other names
3-Dehydroshikimate 3-DHS (−)-3-DHS | |
| Identifiers | |
| 3D model (Jmol) | Interactive image |
| ChEBI | CHEBI:30918 |
| ChemSpider | 388830 |
| PubChem | 439774 |
| |
| |
| Properties | |
| C7H8O5 | |
| Molar mass | 172.14 g·mol−1 |
| Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
3-Dehydroshikimic acid is a chemical compound related to shikimic acid. 3-DHS is available in large quantity through engineering of the shikimic acid pathway.[1]
Metabolism
Biosynthesis: The enzyme 3-dehydroquinate dehydratase uses 3-dehydroquinate to produce 3-dehydroshikimate and H2O.
3-Dehydroshikimate is then reduced to shikimic acid by the enzyme shikimate dehydrogenase, which uses nicotinamide adenine dinucleotide phosphate (NADPH) as a cofactor.
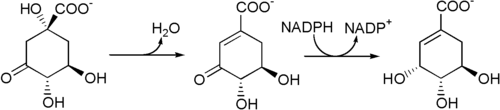 Biosynthesis of shikimic acid from 3-dehydroquinate
Biosynthesis of shikimic acid from 3-dehydroquinate
Gallic acid is also formed from 3-dehydroshikimate by the action of the enzyme shikimate dehydrogenase to produce 3,5-didehydroshikimate. This latter compound spontaneously rearranges to gallic acid.[2][3][4]
References
- ↑ Banwell, M. G.; Edwards, A. J.; Essers, M.; Jolliffe, K. A. (2003). "Conversion of (−)-3-Dehydroshikimic Acid into Derivatives of the (+)-Enantiomer". The Journal of Organic Chemistry. 68 (17): 6839–6841. doi:10.1021/jo034689c. PMID 12919063.
- ↑ Gallic acid pathway on metacyc.org
- ↑ Dewick, P. M.; Haslam, E. (1969). "Phenol biosynthesis in higher plants. Gallic acid". The Biochemical Journal. 113 (3): 537–542. doi:10.1042/bj1130537. PMC 1184696
 . PMID 5807212.
. PMID 5807212. - ↑ Muir, R. M.; Ibáñez, A. M.; Uratsu, S. L.; Ingham, E. S.; Leslie, C. A.; McGranahan, G. H.; Batra, N.; Goyal, S.; Joseph, J.; Jemmis, E. D.; Dandekar, A. M. (2011). "Mechanism of gallic acid biosynthesis in bacteria (Escherichia coli) and walnut (Juglans regia)". Plant Molecular Biology. 75 (6): 555–565. doi:10.1007/s11103-011-9739-3. PMC 3057006
 . PMID 21279669.
. PMID 21279669.
This article is issued from Wikipedia - version of the 2/9/2016. The text is available under the Creative Commons Attribution/Share Alike but additional terms may apply for the media files.